1. Obesogenic high-fat diet-induced intestinal epithelium dysfunction links microbiota dysbiosis and non-communicable diseases
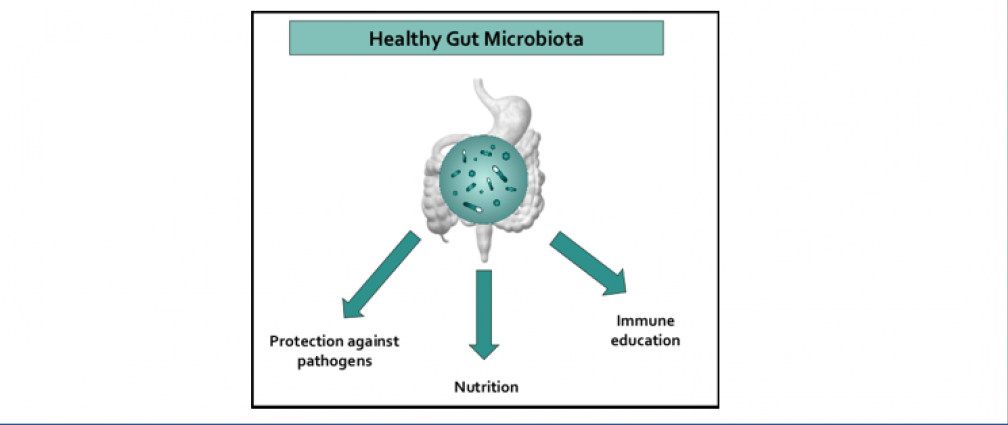
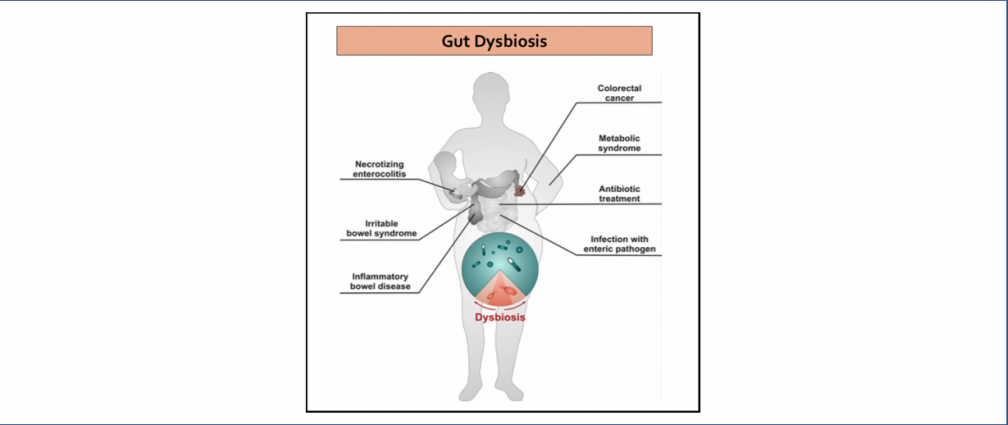
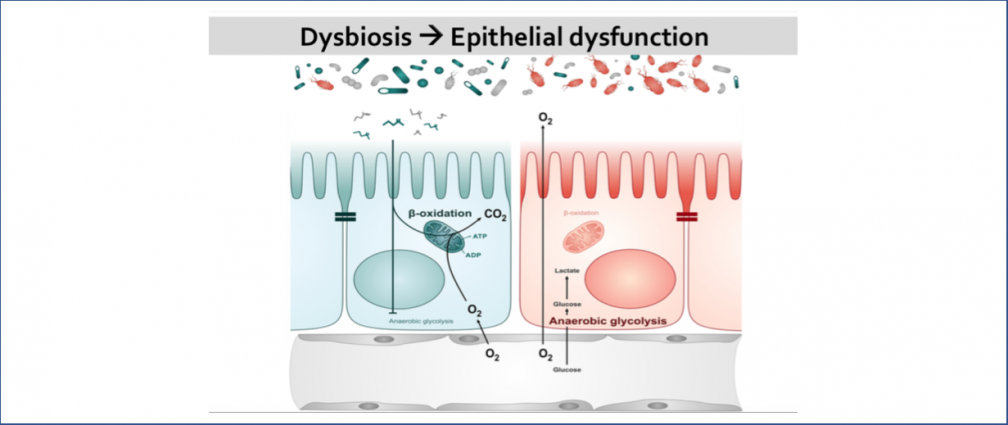
An obesogenic Western-style high-fat (HF) diet causes low-grade intestinal inflammation, intestinal microbiota imbalance, and an increased risk of non-communicable diseases such as cardiovascular disease and colorectal cancer. However, our knowledge about the pathways producing each of these different disease manifestations is incomplete, making it difficult to see connections. Our long-range goal is to elucidate molecular mechanisms that make the gut microbiota a liability during non-communicable diseases. Our research group aims to study the mechanisms through which HF diet drives gut dysbiosis and determine whether the resulting imbalance escalates the production of microbial metabolites that increase the risk for cardiovascular disease and colorectal cancer.
2. Role of microbiota and intestinal epithelial interactions in childhood obesity
Childhood obesity is a disease that affects 20% of U.S. school-age children, leading to an increased risk of chronic health conditions such as type 2 diabetes, cardiovascular disease, and cancer. Early-life microbiota disruption by antibiotics is associated with an increased risk of overweight status in children and mouse models of obesity, particularly those exposed to an obesogenic high-fat (HF) diet. However, our knowledge of the pathways linking antibiotics during childhood, consumption of HF diet, and obesity are incomplete, making it challenging to uncover connections. Our goal is to (i) elucidate a novel mechanism through which antibiotic and HF diet-mediated changes in the intestinal microbiota during early-life affect intestinal epithelial function, (ii) determine how the resulting intestinal epithelial dysfunction drives systemic metabolic changes that lead to obesity, (iii) determine if therapeutically targeting intestinal epithelial function can prevent antibiotic-associated childhood obesity.
3. Mechanisms of Salmonella-mediated disruption of colonization resistance in the inflamed gut
Infection with non-typhoidal Salmonella is 1 of 4 most prevalent global causes of diarrheal disease. In the United States, Salmonella enterica serovar Typhimurium (S. Tm) infection results in 1.35 million illnesses annually. To infect the gastrointestinal tract, S. Tm contends with the resident commensal bacteria (gut microbiota). The gut microbiota benefits the host by limiting enteric pathogen expansion (colonization resistance), partially via the production of inhibitory metabolites such as short-chain fatty acids (SCFA) (e.g., propionate) and nutrient sequestration (e.g., amino acids). Thus, successful bacterial pathogens must possess mechanisms to survive in the competitive ecosystem of the gut. S. Tm disrupts the host-microbiota ecosystem and overcomes microbiota-mediated colonization resistance by using inflammation-derived electron acceptors such as fumarate and nitrate for anaerobic respiration. However, the mechanisms that drive Salmonella-induced disruption of the microbial ecosystem in the gut and how this disruption affects host physiology and promotes pathogen expansion remain largely unknown. Our lab focuses on studying the mechanisms by which S. Tm-induced intestinal inflammation enables the pathogen to (i) overpower SCFA-mediated colonization resistance and (ii) gain access to microbiota-derived amino acids that support S. Tm growth.