Urinary Tract Infections in Cystic Fibrosis Patients
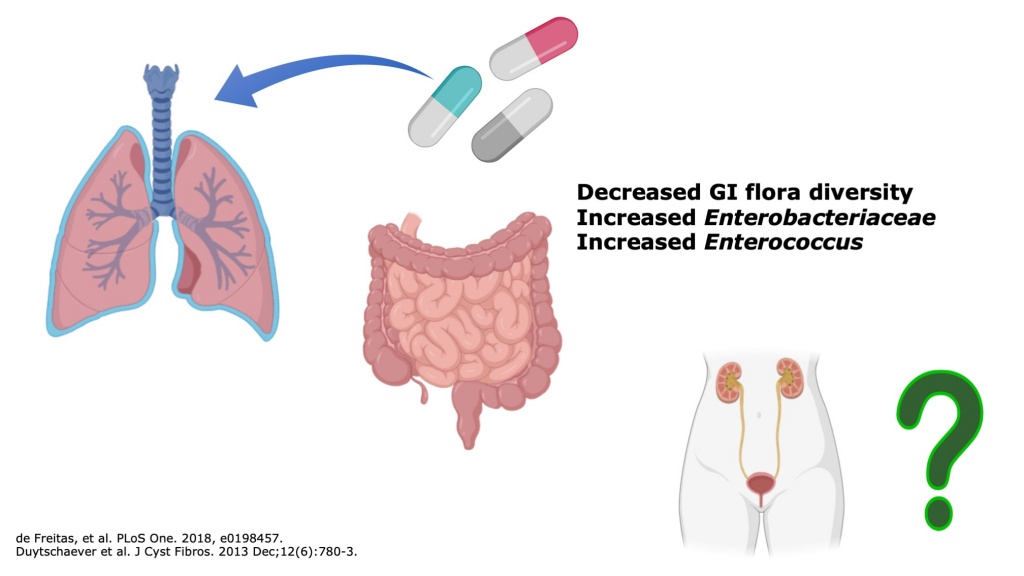
The life expectancy of cystic fibrosis (CF) patients has increased significantly in recent decades but remains limited by recurrent pulmonary infections. CF patients receive frequent antibiotic treatments for respiratory infections. As a result of antibiotic exposure, the gastrointestinal microbiome of CF patients is significantly altered with less susceptible microbes and increased mucosal inflammation.
While cystic fibrosis does not directly affect the urinary system, urinary tract infections (UTI’s) are among the most common bacterial infections and result from translocation of enteric bacteria to the urinary system. To our knowledge, there has not been a published report of the frequency or causative organisms of UTI’s in CF patients. We hypothesized that the altered CF gastrointestinal microbiome may influence the features of UTI’s in CF patients. Using VUMC’s deidentified medical record, the Synthetic Derivative, we intend to perform retrospective chart to determine the frequency and microbial features of UTI’s in CF patients. As the life expectancy of CF patients continues to rise, improved understanding of non-pulmonary infections such as UTI’s will be vital for ensuring longevity and adequate treatment.
Project Personnel:
- Seth Reasoner, MSTP (MD/PhD) student
- Kyle Enriquez, MSTP (MD/PhD) student
- Ben Abelson, MD, Pediatric Urology Fellow
- Michael O’Connor, MD, Pediatric Pulmonologist
- Maria Hadjifrangiskou, PhD, CPMI and microVU Co-Director
Characterizing Clinical Pseudomonas Samples
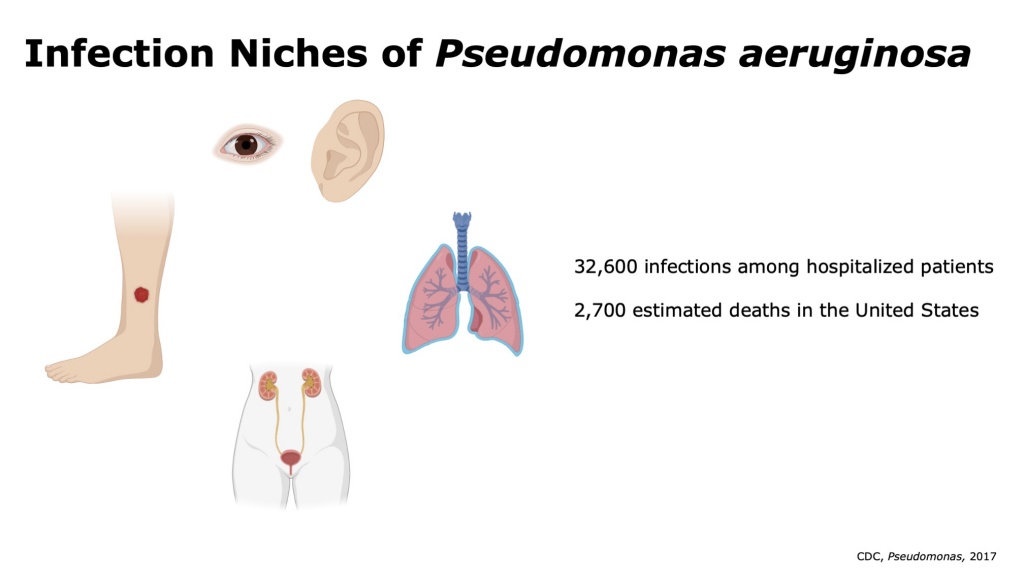
Pseudomonas aeruginosa is a Gram-negative opportunistic pathogen that causes infections in hospitalized and immunocompromised patients. Common sites of P. aeruginosa infections include wound infections in diabetic and burn patients, ventilator associated pneumonia, respiratory infections in cystic fibrosis patients, and urinary tract infections. In each of these infection niches, P. aeruginosa faces vastly different nutrient availability, host immune pressures, and competing microbes. Notably, these infections span the spectrum of temporality from acute infections to chronic recurrent infections. P. aeruginosa presents a challenge to clinicians due to high rates of antibiotic resistance. P. aeruginosa is recognized by the World Health Organization as a “priority pathogen” for new antibiotic development given its increasingly limited treatment options and threat to human health.
Using microVU, we intend to store clinical isolates of Pseudomonas from VUMC’s hospitals and clinics. We intend to pair clinical samples with deidentified patient data to facilitate translational research and comparison of P. aeruginosa across infection sites. Importantly, these studies will provide the first large sample size genomic and transcriptomic comparative analysis across P. aeruginosa clinical infection niches.
Project Personnel:
- Seth Reasoner, MSTP (MD/PhD) student
- Gerald Van’Horn, PhD, D(ABMM)
- Maria Hadjifrangiskou, PhD, CPMI and microVU Co-Director
The Microbiome in Early-Onset Colorectal Cancer
The rate of early-onset colorectal cancer (diagnosis <45 years old) has increased markedly in recent decades for reasons that remain unclear. An increasing body of evidence has demonstrated that alterations in the gastrointestinal microbiome parallel gastrointestinal disease, including Inflammatory Bowel Disease and colorectal cancer, through a common pathway of intestinal inflammation. Leveraging Vanderbilt’s Center for Personalized Microbiology in partnership with VICC’s Young Adult Cancers Initiative, we aim to characterize the microbiome of patients with early-onset colorectal cancer. Stool samples will be collected for metagenomic sequencing to identify possible changes in the gastrointestinal microbiome that may influence the susceptibility to the development of colorectal cancer.
Project Personnel:
- Mariana Byndloss, DVM/PhD, Assistant Professor of Pathology, Microbiology and Immunology
- Cathy Eng, MD, Director of VICC Young Adult Cancers Initiative
- Maria Hadjifrangiskou, PhD, CPMI and microVU Co-Director
Distinguishing a molecular signature for asymptomatic bacteriuria
The objective of this project is to define a molecular signature for asymptomatic bacteriuria (ASB), while illuminating bacterial features that contribute to pathogenesis. Urinary tract infections (UTI) are among the most common infections afflicting humans, with Escherichia coli remaining the primary culprit of the disease. A fundamental gap exists in understanding why some strains of E. coli cause acute infection, while others merely colonize the bladder without apparent symptoms in the common condition of asymptomatic bacteriuria (ASB). The inability to readily discern asymptomatic from symptom-causing urinary isolates currently hampers proper healthcare, leading to over-prescription of antibiotics and contributing to adverse results like gut dysbiosis and selection for antibiotic resistance. Recent work from our groups demonstrated that ASB strains appear to salvage hypoxanthine during growth in human urine. However, in animal models, pathogenic strains of E. coli must synthesize purines de novo to survive within bladder epithelial cells. These results indicate that a potential distinction between ASB and UPEC isolates lies in the way they meet purine requirements in the host. We hypothesize that de novo purine synthesis allows pathogenic E. coli to overcome DNA damage caused by the oxidative stress experienced in the host, while ASB isolates circumvent this need. Further, we hypothesize that a molecular signature that distinguishes ASB exists, but it lies in subtle genomic differences that alter the purine metabolism properties of ASB strains. In two complementary yet independent aims, we will identify the bacterial genomic and metagenomic signatures that drive differential use of the purine biosynthesis pathway in ASB and UPEC strains (Aim 1) and will determine the host-dependent determinants of bacterial purine metabolism during acute UTI and ASB (Aim 2).
Project Personnel:
- Grace Morales, graduate student
- Ben Abelson, MD, Pediatric Urology Fellow
- Jonathan E. Schmitz, MD, PhD, CPMI and microVU Co-Director
- Maria Hadjifrangiskou, PhD, CPMI and microVU Co-Director
Defining microbiome correlations with benign urologic disease across the lifespan
Non-cancerous (i.e. “benign”) genitourinary conditions, such as urinary stone disease (USD) and overactive bladder (OAB), are very common in U.S. children and adults. Recent attention has focused on the urinary microbiome as a novel opportunity to examine microbe-host interactions in urinary diseases, with hope of uncovering pathophysiologic, diagnostic, and/or prognostic information that can help manage these conditions. While prior research has been highly successful at uncovering the nascent role of urinary microbiome in many of these conditions, a systematic, large-scale and transdisciplinary approach to studying the genitourinary microbiome is needed to translate an understanding of urinary microbiome signatures into usable clinical information for decision-making, especially across the life span (i.e. childhood, adolescence, and young, middle, and older adulthood) and between sexes. The overall objective of this application is to define and compare characteristics of the urinary microbiome for individuals with and without USD and OAB identified through cutting- edge, culture-independent techniques (i.e. 16S RNA and metabolomic sequencing), which may be used as diagnostic and prognostic clinical indicators.
Project Personnel:
- Douglass Clayton, MD, Pediatric Urologic Surgery
- W. Stuart Reynolds, MD, Adult Urology
- Ryan His, MD, Adult Urology